The Digital Pathology Illusion: Why Are We Building Storage Instead of Solutions?

by Rajendra Singh MD
Professor of Pathology, University of Pennsylvania
Co-Founder, PathPresenter
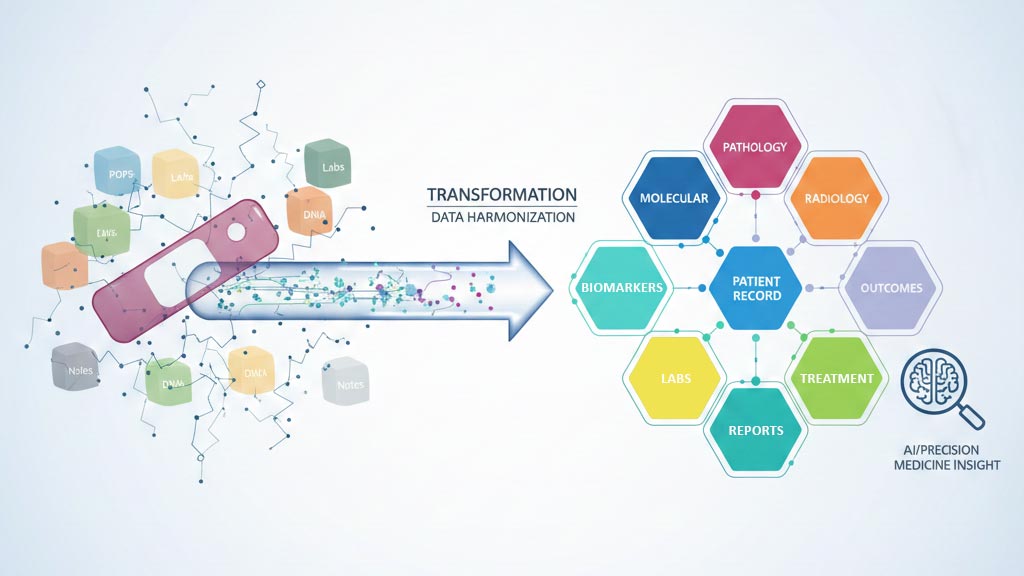
Digital pathology has finally moved from “nice-to-have” to a global strategic priority. The benefits are real and include remote sign-out, operational efficiency, collaboration, education, and the promise of AI. But as health systems rush to digitize, many are falling for a dangerous misconception: that digital slides automatically equal a valuable dataset.
They don’t.
Right now, we’re witnessing a massive and expensive race to digitize pathology, yet we’re often solving the wrong problem. If your organization’s primary focus is “where to store the pixels,” you’re not building a future for AI, on the contrary, you’re building a more expensive filing cabinet.
The truth: no one wants your slides
It’s time for a reality check: no researcher, pharma partner, or AI developer is looking for “just digital slides.” A WSI in isolation is a clinical dead end and actually a financial liability. Every day that pixel data sits in a silo without clinical context, it consumes storage costs while generating zero insights. A slide only becomes valuable when it’s connected to the patient’s clinical truth and metadata that link to tumor type and subtype, molecular and prognostic markers, treatment course, and outcomes (response, recurrence/progression, survival). Without that context, a WSI is often just a beautiful high-resolution image file with limited real-world utility.
We digitized the workflow, not the intelligence.
Most institutions today have a “digital pathology” environment that is still a graveyard of silos: slides live in scanner storage or cloud buckets, reports sit elsewhere in the LIS/EMR as static PDFs, and molecular results, labs, treatments, and outcomes remain buried in separate systems or unstructured clinical notes. Even within pathology, the report is still largely narrative text, so downstream “metadata” often stops at age, sex, and diagnosis, while key details like biomarkers, margins, tumor size, staging, and prognostic factors remain trapped in free text. In short: we’ve digitized pathology, but we haven’t structured it, and without structure, we don’t have a dataset, we have digital clutter.
The real dataset is a unified patient story
Real value of the kind that accelerates research, supports precision medicine, and enables reliable AI only exists when data is multi-modal, longitudinal, and semantically organized. That means slides are programmatically linked to pathology findings, biomarkers/molecular data, labs, radiology, treatment events, and outcomes, following the patient journey over time. Researchers don’t search for “tumor” vs “cancer” vs “malignancy”. They search by clinical meaning: triple-negative breast cancer, EGFR-mutated lung adenocarcinoma on targeted therapy, high-risk disease with recurrence. That requires semantic organization, not just storage.
And this is becoming urgent. We’re entering the era of tabular foundation models, where AI learns from structured clinical tables at scale. These models are only as good as the structure and consistency of the data they ingest. Messy, disconnected silos don’t just slow AI down, they undermine its reliability.
The bottom line
“The goldmine of pathology data” will remain a hollow promise until we stop treating the image as the final product. The image is the starting line. The dataset is the unified, searchable, longitudinal record of the patient’s life.
Stop asking: “Where do we store the slides?” Start asking: “How do we build a structured data foundation that connects the pixels to the patient, automatically and at scale?”
If you aren’t solving for data structure, you aren’t solving the problem.
Pathologists are not just the users of these systems; they are the architects of the clinical truth. If the pathologist doesn’t define the structure at the point of capture, the data is lost to the “narrative abyss” of a PDF forever. But realizing its full translational value requires partnership with strong technical teams who understand structured data design, concept libraries and standardized ontologies, flexible database architectures, tabular foundation models, and semantic search. Together, we can turn stored pixels into the most valuable data in healthcare that powers trustworthy AI and ensures every patient receives the right treatment at the right time..
About the Author
Dr. Rajendra Singh is a Professor of Pathology at the University of Pennsylvania and co-founder of PathPresenter. He serves as a member of the Digital and Computational Pathology Committee of the CAP, Editorial Board of the WHO for Classification of tumors, 5th Edition and the Board of Digital Pathology Association.
More Posts
- The Digital Pathology Illusion: Why Are We Building Storage Instead of Solutions?
- Success Story: Accelerating Clinical Trials for Oncology with Imagenomix Predict and PathPresenter ConsultConnect
- Why Seamless IMS-LIS Integration Is Essential for Digital Pathology
- Digital Pathology Basics: Image Viewers vs. Image Management Systems
- Choosing the Right Digital Pathology Workflow Partner: So Many Options
